Optimizing K-mer Counting and Querying in Commodity Clusters
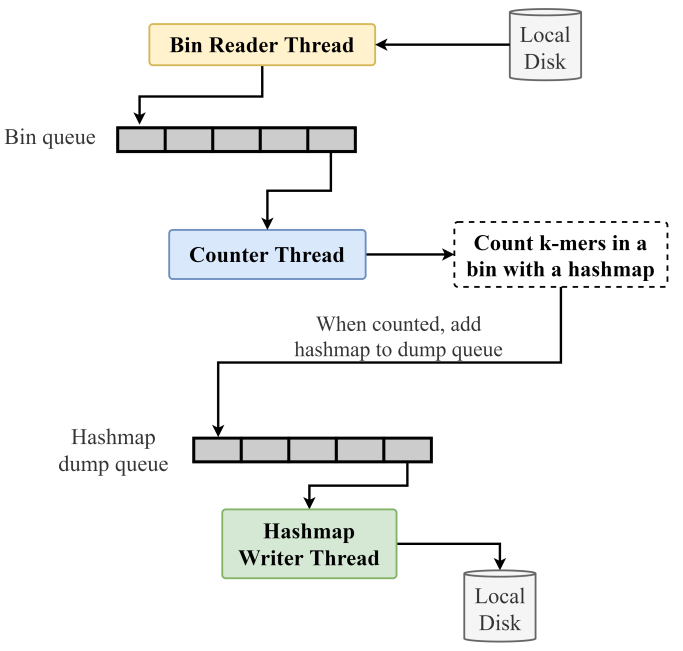
k-mer counting is the process of counting k length substrings in a sequence. It is an important step in many bioinformatics applications including genome assembly, sequence error correction, and sequence alignment. Even though generating k-mer histograms seems simple and straightforward, processing large datasets efficiently with limited resources, especially memory, is very challenging. As the advancements in next-generation sequencing technologies have resulted in a tremendous growth of genomic data, it is inevitable for k-mer counters to be faster and more efficient. A lot of work has been done in the past decade to optimize k-mer counting.
The existing distributed memory solutions are specialized for high performance clusters and high speed networks which are costly to implement. Hence, the focus of this research is to build a k-mer counting and querying tool which is optimized for commodity clusters. We present Quill: a memory-efficient k-mer counting and a querying tool for commodity clusters. Quill manages to perform k-mer counting in a conventional computer cluster without relying on high-performance network interfaces or parallel file systems. Furthermore, Quill provides an additional advantage in cases where k-mer counting is required for multiple k-values in the same dataset.
Quill shows a linear scaling for the k-mer counting stage when tested in a commodity cluster. The performance gain is more evident when executed with k values up to 22 and 28. Thus, Quill can be viewed as a cost-effective k-mer counting solution that can effectively use the combined computing power of a cluster of commodity-grade computers.
Quill is freely available at https://github.com/CSE-Optimizers/k-mer_counter .
Publications
- Edippuliarachchi, B., Gamlath, D., Amaratunga, R., Brihadiswaran, G., & Jayasena, S. (2022). Quill: A Memory Efficient k-mer Counting and k-mer Querying Tool for Commodity Clusters. In Proceedings of the 14th International Conference on Bioinformatics and Biomedical Technology (ICBBT ’22) (pp. 79-88).
